News
Article
AACR 2025: Off-the-Shelf Machine Learning Models May Bridge Diagnostic Gaps in Global Skin Cancer Care
Author(s):
Key Takeaways
- Pretrained foundation models improve NMSC diagnostic accuracy, outperforming traditional methods like ResNet18, especially in resource-limited settings.
- Simplified versions of foundation models maintain high accuracy, making them more accessible for deployment in environments with limited resources.
Pretrained foundation machine learning models significantly improved the accuracy of diagnosing non-melanoma skin cancer from digital pathology images and may offer a practical, resource-efficient solution for cancer diagnosis in underserved settings.
Pretrained machine learning models could play a significant role in diagnosing non-melanoma skin cancer (NMSC) in regions with limited access to expert pathology services. At the 2025 American Association for Cancer Research (AACR) Annual Meeting in Chicago, Illinois, Steven Song, an MD/PhD candidate at the University of Chicago, explained that by harnessing the power of foundation models—large-scale, general-purpose machine learning models trained on extensive datasets—his research team demonstrated improved diagnostic accuracy compared to conventional approaches, particularly in resource-constrained environments.
“In resource-limited settings, the lack of expert pathologists limits the ability to provide timely and widespread review and diagnosis of NMSC,” Song said in an AACR statement. “Artificial intelligence and machine learning have long promised to fill resource gaps, but the development and deployment of bespoke machine learning models require significant resources that may not be available in many places—namely computational experts, specialized computational hardware, and large amounts of curated data to train each model.”
To address this bottleneck in the diagnostic process, Song and his colleagues explored whether pretrained models—often referred to as foundation models—could be repurposed in an “off the shelf” fashion to assist with NMSC diagnosis. According to Song, these models have been trained on vast amounts of data and are designed to generalize efficiently across different domains. Their use eliminates the need to develop new models from scratch for each clinical application, thereby lowering the barriers to deploying AI in settings with limited technical infrastructure.
In their study, the researchers evaluated the performance of 3 prominent foundation models—PRISM, UNI, and Prov-GigaPath—against that of ResNet18, a widely used image recognition architecture, on digital pathology slides of skin tissue from the Bangladesh Vitamin E and Selenium Trial (BEST). According to Song, his team chose slides from the BEST study cohort due to the high prevalence of NMSC in Bangladesh, largely driven by chronic exposure to arsenic-contaminated drinking water.
Skin biopsy pathology of basal cell carcinoma invading the dermis. Image Credit: © David A Litman - stock.adobe.com
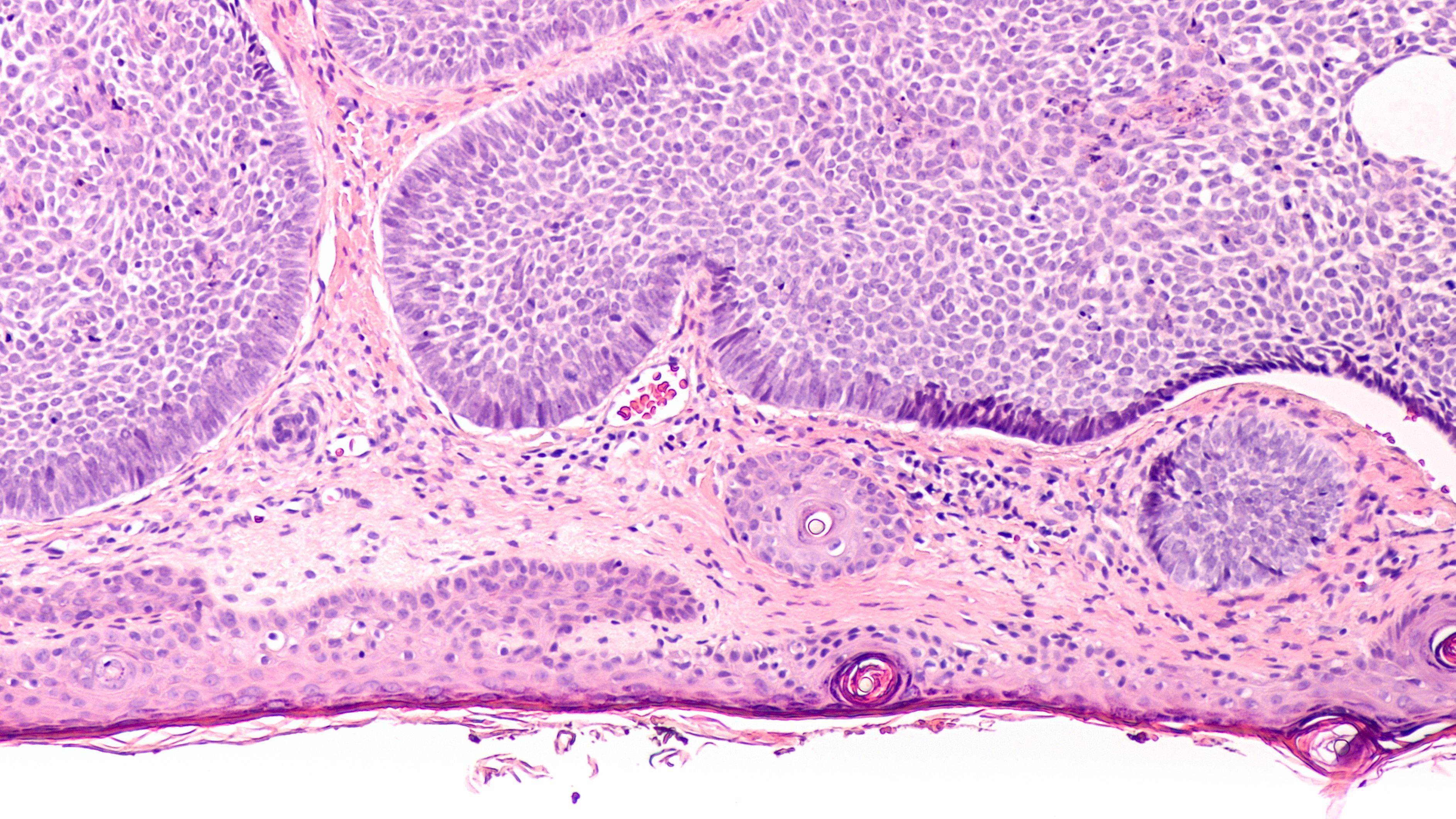
The dataset included 2130 high-resolution digital images from 553 biopsy samples. Of these, 1424 images represented various NMSC types—Bowen’s disease, basal cell carcinoma, and invasive squamous cell carcinoma—while 706 images were of normal tissue. Each foundation model analyzed the tissue slides by breaking them into smaller image tiles, extracting relevant features, and assessing the likelihood of cancerous tissue presence.
The foundation models demonstrated strong performance, significantly surpassing that of ResNet18. ResNet18 achieved an accuracy of 80.5% in distinguishing between cancerous and non-cancerous tissue. In contrast, PRISM achieved an accuracy of 92.5%, UNI 91.3%, and Prov-GigaPath 90.8%.
These results underscore the power of pretrained models in pathology. However, Song’s team also recognized that even these models, while more accessible than training a new one from scratch, may still be too complex or resource-intensive for some environments. To make deployment more feasible, they developed simplified versions of each foundation model that required less data processing. Encouragingly, these streamlined versions still performed well, achieving accuracies of 88.2% (PRISM), 86.5% (UNI), and 85.5% (Prov-GigaPath), again outperforming the ResNet18 baseline.
Beyond classification accuracy, the team also introduced a method for annotating cancerous regions on digital slides. This framework does not require extensive training data; instead, it uses a few annotated examples to help highlight potentially cancerous regions in new images. This kind of visual aid could prove extremely useful in guiding clinicians or technicians who may not be trained pathologists but still play a role in the diagnostic process.
From a technical perspective, the study employed a rigorous evaluation framework, using 5-fold cross-validation and comparing various combinations of embedding strategies and classifiers. For instance, the best performing model used PRISM embeddings aggregated through its intrinsic Perceiver network and classified with a shallow multilayer perceptron, achieving a mean area under the receiver operating characteristic curve of 0.925. Even simple classifiers such as logistic regression performed well when combined with global average pooling of foundation model embeddings, suggesting flexibility in adapting the system to available computational resources.
Despite these promising results, the authors acknowledged limitations. Most notably, the study evaluated the models on a single population of Bangladeshi individuals. While this cohort was well-suited for studying NMSC due to high disease prevalence in this population, the findings may not be fully generalizable to other populations with different genetic, environmental, or health care characteristics. Additionally, although the study was motivated by the needs of resource-limited settings, it did not directly address logistical challenges of real-world deployment, such as the availability of slide-scanning hardware, internet connectivity, integration into existing clinical workflows, and the need for user training.
Even with these caveats, the work highlights a practical pathway toward narrowing global health disparities in cancer care. By leveraging pretrained machine learning models, it may become possible to extend high-quality diagnostic tools to communities that lack access to pathologists. The research also points to the broader potential of foundation models in medicine, where a single robust model can be adapted for various tasks with relatively minimal customization.
Looking ahead, further studies will be needed to validate these models across diverse settings and to pilot real-world implementation. Questions remain about the sustainability of this approach with the need for maintenance and updates of the AI tools, as well as how to best ensure data privacy and ethical use of patient images.
“While our study suggests foundation models as resource-efficient tools for aiding NMSC diagnosis, we acknowledge that we are still far from having a direct impact on patient care,” Song said in the AACR statement. “Further work is needed to address practical considerations, but the potential is real—and promising.”
REFERENCE
Pretrained Machine Learning Models May Help Accurately Diagnose Nonmelanoma Skin Cancer in Resource-limited Settings. American Association for Cancer. April 28, 2025. Accessed April 25, 2025. https://aacr.ent.box.com/s/sm6x7i8w4l87qfhcrjtl877wodplhtrg
Newsletter
Stay informed on drug updates, treatment guidelines, and pharmacy practice trends—subscribe to Pharmacy Times for weekly clinical insights.